Download
Installers are available for Windows, MacOS and several Unix distro's. PPAs are available Debian and Ubuntu and AUR packages for ArchLinux.
User Guide
Get the lattest documentation on the full features of TriFusion, how to operate GUI and command line versions of TriFusion, and more!
Download guideView on GitHub
TriFusion is and will always be free software (under GPLv3 license). Feel free to view and modify the source code, hosted on GitHub.
View SourceHandling phylogenomic data is much easier now.
TriFusion was specifically developed to streamline several stages of phylogenomic and population genomic projects, from the gathering of orthologs from hundreds of proteomes, to the processing of thousands of alignment files, to data exploration via an assortment of graphical and statistical analyses.
See full features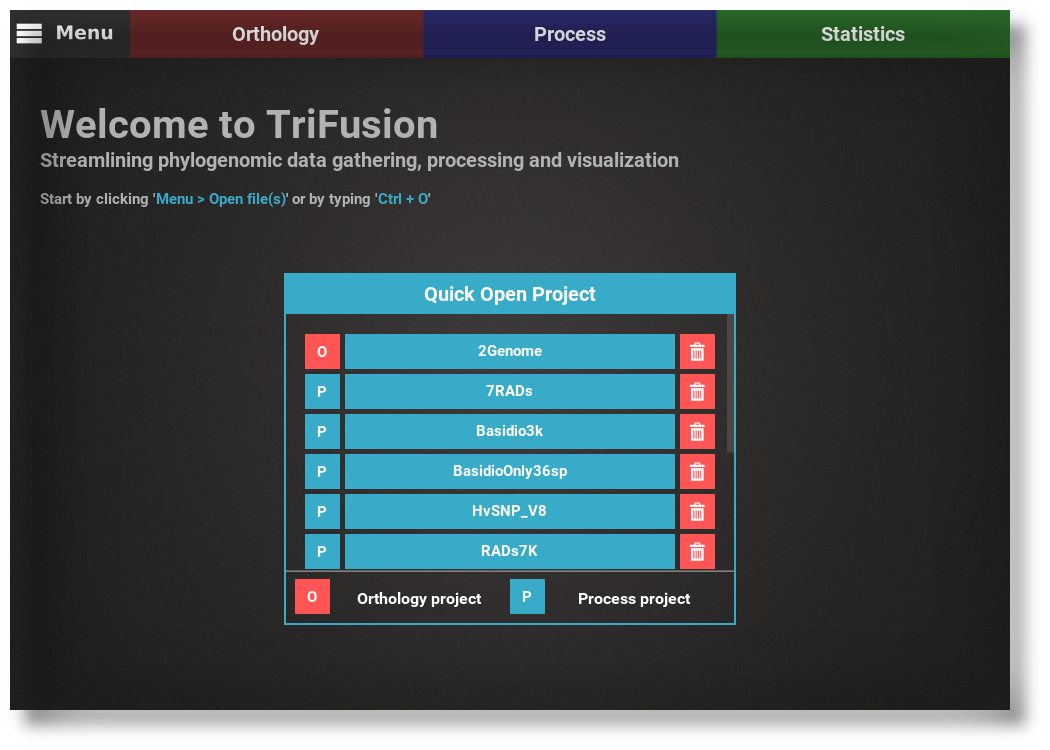
Gather and explore orthologs from multiple proteomes with just a few clicks.
Load proteome files in Fasta format from hundreds of species into TriFusion and get the full coverage of high quality ortholog clusters with minimal user input. Take advantage of several filters and graphical options to explore your ortholog groups and export them into protein and/or nucleotide sequences.
See Orthology features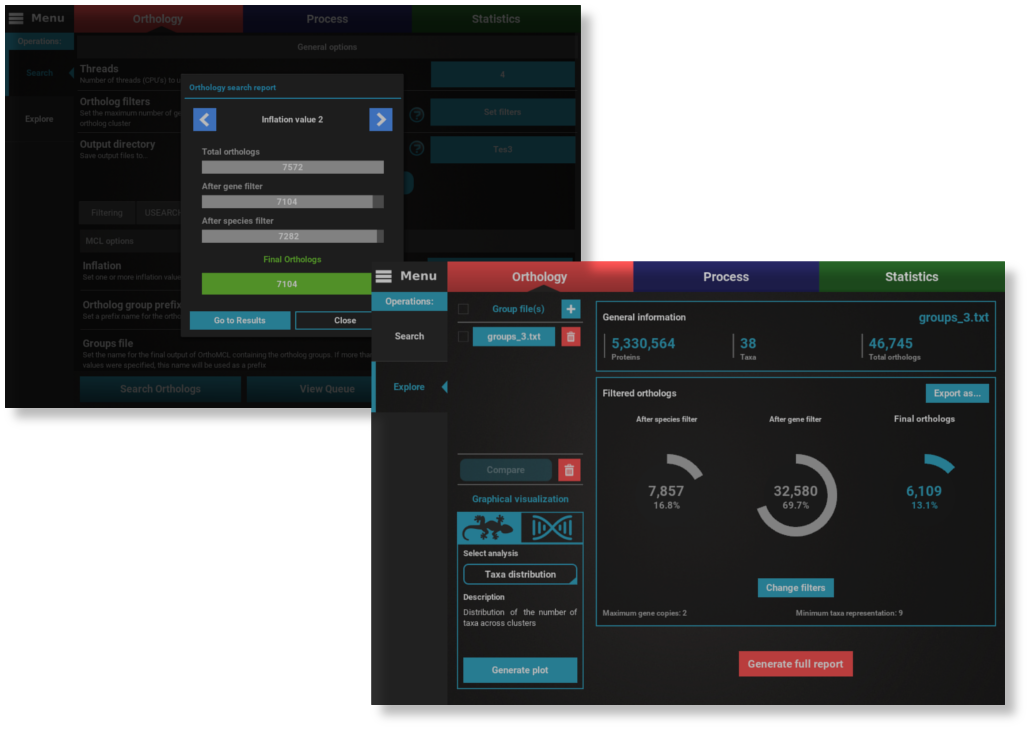
Convert, concatenate, filter, collapse alignments and so much more!
TriFusion offers a wide variety of useful and common operations that can be quickly and easily applied to thousands of alignment files. Want to collapse identical sequences into haplotypes? Check! Want to filter alignments with excess of missing data? Check! Want to remove certain codon positions? Check! Want to create a consensus from multiple sequences in an alignments? Check! See the user manual for the complete set of operations that you can do.
See Process features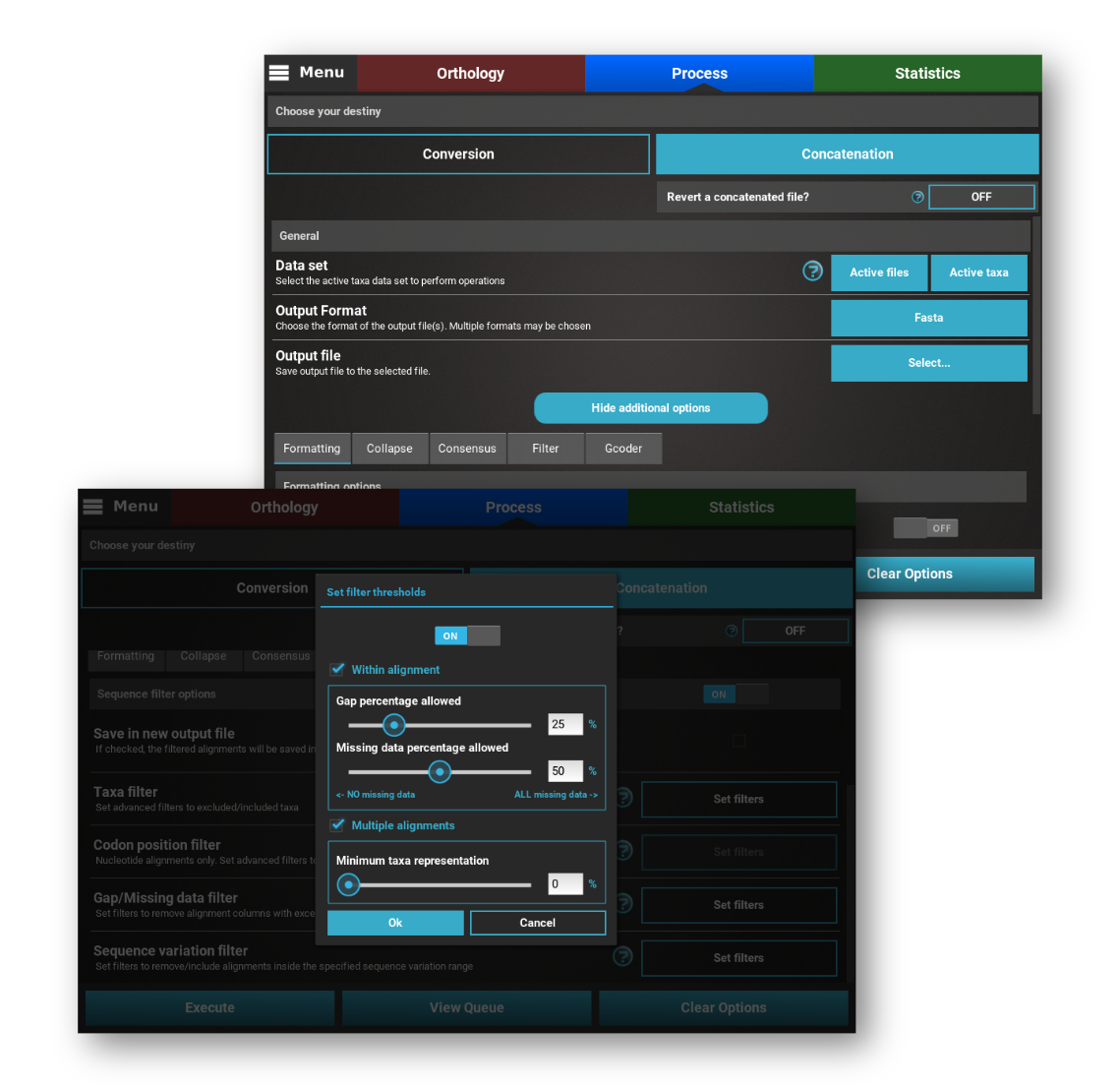
Full graphical visualization of your data
TriFusion tackles one of the biggest challenges in big phylogenomic and population genomics projects: How to explore your data quickly and efficiently. Designed from the start to handle thousands of alignment files, the Statistics module offers dozens of statistical and graphical options to explore your data according to four main categories: General Information, Polymorphism and Variation, Missing Data and Outlier Dectection
See Statistics features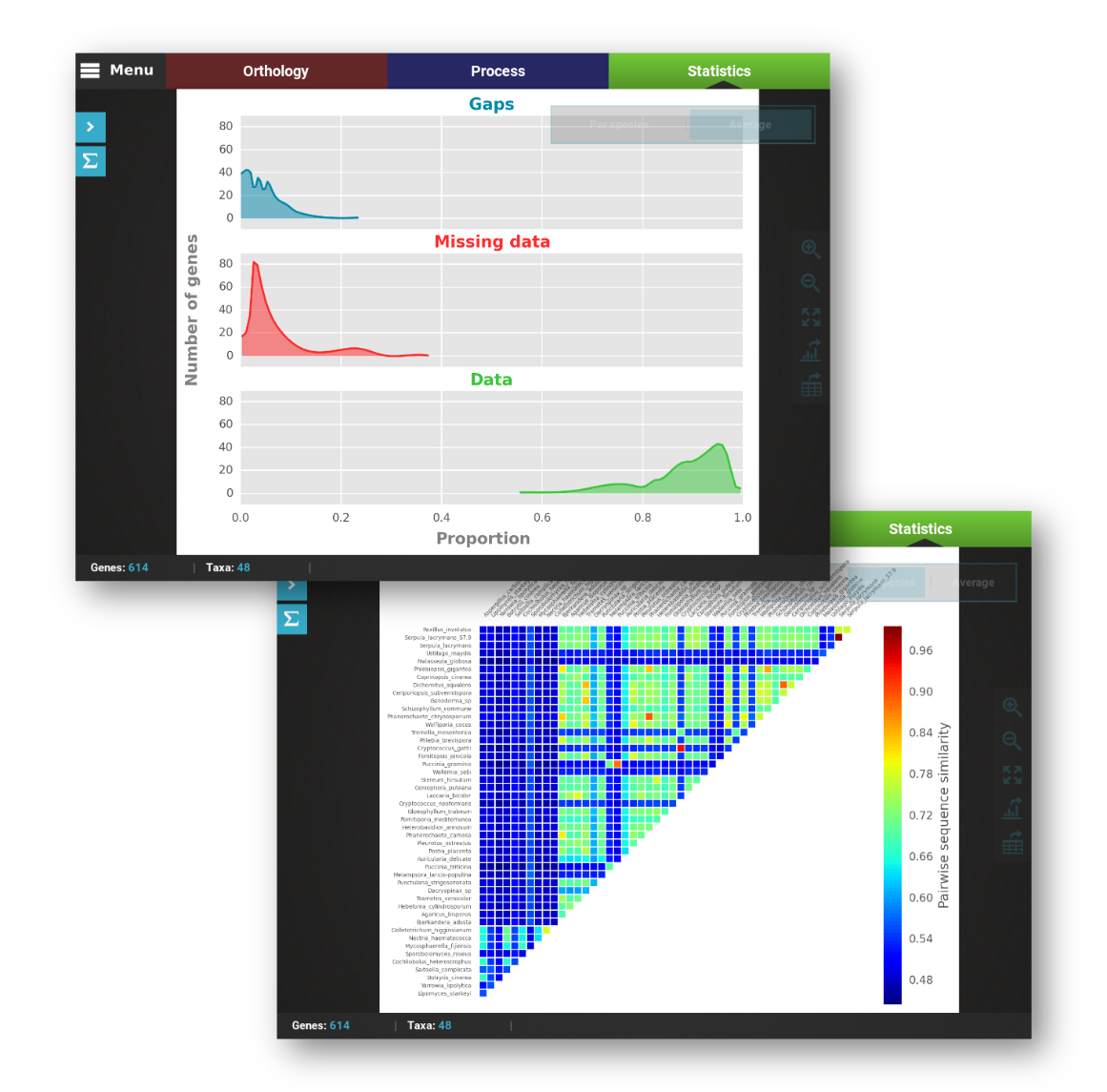
For command line lovers <3
Almost all functionalities of TriFusion can be executed from a terminal by using the appropriate CLI programs: orthomcl_pipeline (Orthology Search), TriSeq (Process) and TriStats (Statistics). The CLI programs are faster, modular and can be easily included in your scripts. This also means that TriFusion can be executed in "GUI-free" servers for tasks that may require high uptimes and/or computational resources.

Jump right in into TriFusion using any of the following quick start guides. These are sorted according to TriFusion's three main modules:
General
- The three ways to load data into TriFusion.
- Creating and using active data set groups.
- Projects for quick data loading.
Orthology
- Setup of USEARCH.
- Basic search of orthologs among 10 proteome files.
- Import and explore multiple orthology search results.
- Export ortholog groups into protein and nucleotide sequence files.
Process
- Limitations for input files
- Basic conversion and concatenation of multiple alignment files.
- Reverting a concatenated file.
- Applying secondary operations (collapse, consensus, filtering, gap coding).
- Working with partitions and substitution models




